This article has been reviewed according to Science X's editorial process and policies. Editors have highlighted the following attributes while ensuring the content's credibility:
fact-checked
peer-reviewed publication
proofread
Using cancer proteomics data to identify gene candidates for therapeutic targeting
A new research perspective titled "Using cancer proteomics data to identify gene candidates for therapeutic targeting" has been published in Oncotarget.
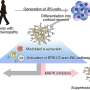
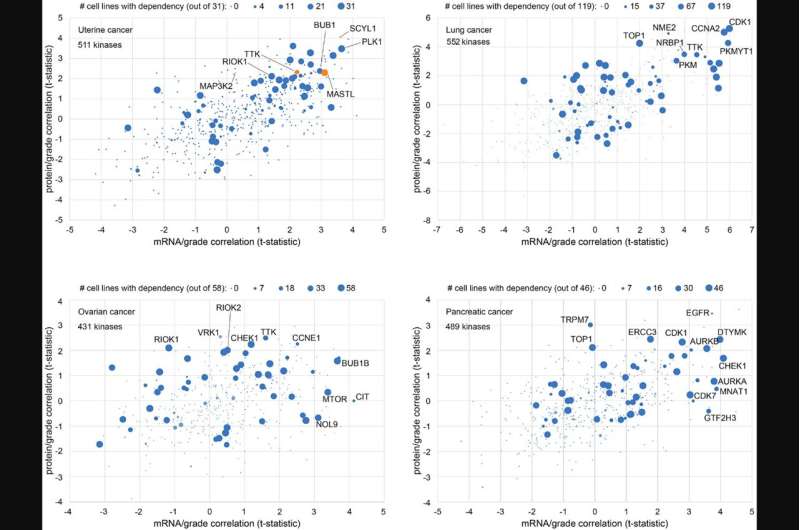
Gene-level associations obtained from mass-spectrometry-based cancer proteomics datasets represent a resource for identifying gene candidates for functional studies. In their new research perspective, researchers Diana Monsivais, Sydney E. Parks, Darshan S. Chandrashekar, Sooryanarayana Varambally, and Chad J. Creighton from Baylor College of Medicine and the University of Alabama at Birmingham discuss their recent study where they surveyed proteomic correlates of tumor grade across multiple cancer types and identified specific protein kinases having a functional impact on uterine endometrial cancer cells.
"This previously published study provides just one template for utilizing public molecular datasets to discover potential novel therapeutic targets and approaches for cancer patients," the researchers explain.
Proteomic profiling data combined with corresponding multi-omics data on human tumors and cell lines can be analyzed in various ways to prioritize genes of interest for interrogating biology. Across hundreds of cancer cell lines, CRISPR loss of function and drug sensitivity scoring can be readily integrated with protein data to predict any gene's functional impact before bench experiments are carried out. Public data portals make cancer proteomics data more accessible to the research community. Drug discovery platforms can screen hundreds of millions of small molecule inhibitors for those that target a gene or pathway of interest.
"Here, we discuss some of the available public genomic and proteomic resources while considering approaches to how these could be leveraged for molecular biology insights or drug discovery. We also demonstrate the inhibitory effect of BAY1217389, a TTK inhibitor recently tested in a Phase I clinical trial for the treatment of solid tumors, on uterine cancer cell line viability," note the researchers.
More information: Diana Monsivais et al, Using cancer proteomics data to identify gene candidates for therapeutic targeting, Oncotarget (2023). DOI: 10.18632/oncotarget.28420