LA JOLLA, CA—A new study led by scientists at La Jolla Institute for Immunology (LJI) shows how ideal antibodies against SARS-CoV-2 hit their marks.
That’s right: marks. Instead of targeting a single binding site on the SARS-CoV-2 Spike protein, these powerful antibodies bind to two of these sites at once. Through this “bivalent” binding, the antibodies can lock key viral structures into position, preventing the virus from infecting host cells.
The new findings, published in Cell Reports, suggest bivalent antibodies retain efficacy against both early SARS-CoV-2 variants and several later Omicron variants. Now scientists are looking at how we might harness their power in new antibody therapeutics and even more effective COVID-19 vaccines.
“The ideal antibodies exist,” says Kathryn Hastie, Ph.D., an LJI Instructor and the Director of the Antibody Discovery Center at LJI. Hastie led the new study alongside LJI Postdoctoral Researcher Heather Callaway, Ph.D., Sharon Schendel, Ph.D., and LJI President and CEO Erica Ollmann Saphire Ph.D. “Now the question is, how do we preferentially boost those?”
The new research was made possible through the Coronavirus Immunotherapy Consortium (CoVIC), a global effort with Saphire as Director and Schendel as Program Manager.
How well do your antibodies target Omicron and other SARS-CoV-2variants? It depends on their structure. LJI researchers summarize here the many ways antibodies target the Omicron receptor binding domain (RBD). (Credit: Saphire Lab, LJI))
Antibodies vs. Omicron
Current COVID-19 vaccines are designed to teach the body to recognize the SARS-CoV-2 Spike protein. Spike is important because it recognizes and latches onto host cells through a region called the receptor binding domain (RBD). Each spike has three copies of the RBD. An antibody that can bind to a target (called an epitope) on the RBD has a chance to jam up this machinery and stop infection.
[Learn more: From Alpha to Epsilon: Consortium study illuminates surfaces of Spike most resistant to antibody escape]
The problem is that SARS-CoV-2 keeps mutating, and many once-powerful antibodies can’t recognize their usual targets on RBDs. Building vaccines and therapies that will withstand future variation both hinge on figuring out what surviving antibodies have in common.
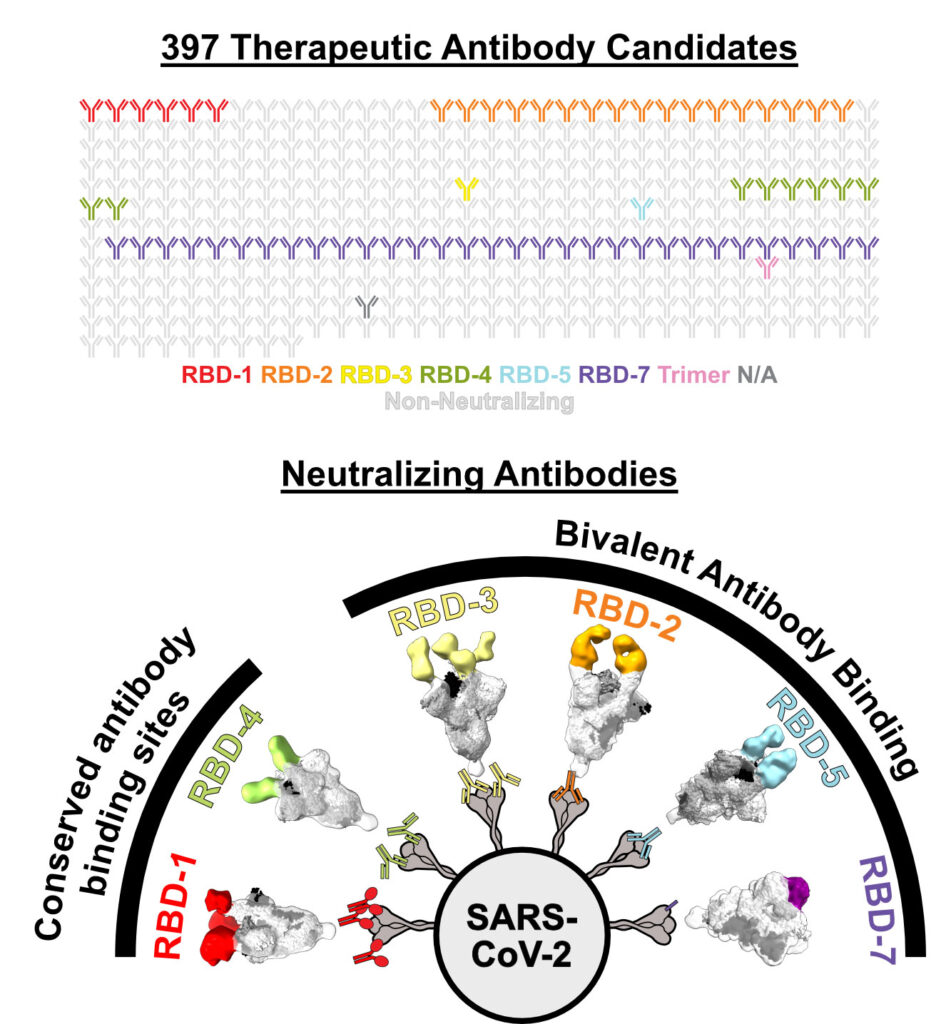
Callaway and Hastie analyzed nearly 400 antibodies sent by scientists around the world to the CoVIC, which is headquartered at LJI. They narrowed down this huge pool and found 66 potent antibodies that could neutralize Omicron lineage BA.1 and early sub-lineages such as BA.1.1 and BA.2. By the time they tested antibodies against Omicron lineages BA.4/5, only seven antibodies had neutralizing power.
The issue was, these surviving antibodies targeted the same spot as other antibodies that didn’t withstand the mutations in Omicron. It was time to see how this unique group of neutralizing antibodies was getting the job done. Callaway used an imaging technique called cryo-electron microscopy and biochemical analysis to see exactly how these antibodies bound to molecules in the RBD.
“It became clear there was a pattern,” says Callaway. “The successful antibodies overcome loss of affinity for Omicron by simultaneously binding two RBDs in the Spike.”
The best neutralizing antibodies hung on with both hands. They didn’t bind their targets, one at a time, especially strongly, but by locking together two adjacent RBDs at once, even Omicron had a tough time escaping. Bivalent binding equaled antibody survival against the latest variants.
The researchers grouped antibodies into communities (see color-coded groups above) based on precisely where the antibodies target the RBD. The new study highlights how some communities maintain their neutralizing power against Omicron lineage BA.1. (Credit: Saphire Lab, LJI)
A closer look at bivalent antibodies
An antibody’s basic structure resembles the letter Y. The two arms at the top of the Y are called the fragment antigen-binding regions (Fabs). The two Fabs on each antibody have the same molecular structure, and Fabs work like keys seeking a lock (an epitope).
Scientists conducting structural analyses of how antibodies bind proteins often use only the Fab region, and images in textbooks have focused on how single Fabs bind to single sites. The new study suggests that this previous focus can lead to the wrong impressions.
Instead, the LJI team has shown the importance of imaging the intact, complete antibody as it exists in nature, a type of analysis only possible with new and powerful electron microscopes. By doing so, the team could distinguish what survived vs. what fell to the wayside upon emergence of Omicron. Antibodies that use both their arms to anchor two RBDs at once withstood viral evolution.
Going forward, the researchers will continue to test the CoVIC pool of antibodies against new SARS-CoV-2 variants. This information is essential to development of better vaccines – that will elicit immunity that lasts multiple seasons of viral evolution. The Saphire Lab is also engineering such immunogens that will retain broad activity through time.
The power of CoVIC
The very fact that researchers were able to analyze a broad pool of nearly 400 antibodies has been a major step forward for science.
As Saphire explains, the broad scope of the CoVIC antibody panel comprising hundreds of therapeutic candidates contributed by over 60 groups in both corporate and academic settings—combined with side-by-side comparisons using a range of techniques—revealed enough of this rare antibody population to explain what features made them special.
“The scale of the study allowed the team to study enough Omicron ‘survivors’ to find general rules about what made them successful,” says Saphire.
“CoVIC demonstrated that collaboration allowing studies on a larger scale can lead to better insights, ” Schendel adds.
Schendel says that having both the Salt Lake City-based biotech company Carterra and the laboratory of Professor Georgia Tomaras, Ph.D., at Duke University as partner reference labs in the CoVIC was especially important for sorting the different neutralizing antibodies into epitope-binding “communities,” based on where they target SARS-CoV-2.
Learning more about these antibodies can guide creation of next-generation antibody cocktails that are more resistant to ever-emerging SARS-CoV-2 variants.
Going forward, it will be important to know if promising antibodies are competing with each other or complementing each other. The LJI researchers also say it’s critical to look at how new vaccines present the Spike protein to the immune system. The more accurate the immunogen version of Spike is, and the more accurately it presents the arrangement and spacing of the component parts, the more likely it will elicit neutralizing antibodies.
“We really need to think about preserving the fold, geometry and overall appearance of the Spike protein,” Schendel says.
Additional authors of the study, “Bivalent intra-spike binding provides durability against emergent Omicron lineages: Results from a global consortium,” include Haoyang Li, Xiaoying Yu, Jeremy Shek, Tierra Buck, Sean Hui, Dan Bedinger, Camille Troup, S. Moses Dennison, Kan Li, Michael Alpert, Charles C. Bailey, Sharon Benzeno, Jody L. Bonnevier, Jin-Qiu Chen, Charm Chen, Hyeseon Cho, Peter D. Crompton, Vincent Dussupt, Kevin C. Entzminger, Yassine Ezzyat, Jonathan K. Fleming, Nick Geukens, Amy E. Gilbert, Yongjun Guan, Xiaojian Han, Christopher J. Harvey, Julia M. Hatler, Bryan Howie, Chao Hu, Ailong Huang, Maya Imbrechts, Aishun Jin, Nik Kamachi, Gladys Keitany, Mark Klinger, Jay K. Kolls, Shelly J. Krebs, Tingting Li, Feiyan Luo, Toshiaki Maruyama, Michael A. Meehl, Letzibeth Mendez-Rivera, Andrea Musa, C.J. Okumura, Benjamin E.R. Rubin, Aaron K. Sato, Meiying Shen, Anirudh Singh, Shuyi Song, Joshua Tan, Jeffrey M. Trimarchi, Dhruvkumar P. Upadhyay, Yingming Wang, Lei Yu, Tom Z. Yuan, Erik Yusko, Bjoern Peters, and Georgia Tomaras
The study was supported by the Bill & Melinda Gates Foundation (grants INV-0006133 and INV-008612), the National Institutes of Health (grant U19 AI142790-03S1) and the GHR Foundation. Additional funding was provided by the NIH National Institute of Allergy and Infectious Diseases (grants R44AI145491 and R44AI134269); the Guangzhou Municipal Science and Technology Bureau (project #202201020528); the US Department of Defense, Defense Health Agency, through the CARES Act; KU Leuven KOOR ZKD8270; KU Leuven SARS-CoV-2 mAb OF2; KU Leuven C3/20/105; the Emergency Project for COVID-19 from Chongqing Medical
University; and through a cooperative agreement (W81XWH-18-2-0040) between the Medical Research and Development Command of the Army Futures Command in the US Department of the Army.
DOI: 10.1016/j.celrep.2023.112014
###
About La Jolla Institute
The La Jolla Institute for Immunology is dedicated to understanding the intricacies and power of the immune system so that we may apply that knowledge to promote human health and prevent a wide range of diseases. Since its founding in 1988 as an independent, nonprofit research organization, the Institute has made numerous advances leading toward its goal: life without disease. Visit lji.org for more information.



