Highlighting the molecular mechanism underlying pancreatic cancer development
Researchers have visualized the protein binding site of the (pro)renin receptor known to be involved in pancreatic cancer development using an artificial intelligence-based protein structure prediction program. They predicted the 3D shape of this receptor and revealed the presence of hand-shaped grooves, allowing for the binding of multiple proteins.
This study provides the first 3D structural insight into the receptor binding and one-to-many interactions, underpinning the functional versatility of this receptor. These findings will increase our understanding of disease pathogenesis and aid us in exploring novel modalities to treat human diseases, including hypertension and cancer.
The (pro)renin receptor [(P)RR] is a cell membrane-bound protein that was originally identified as a potential regulator of the renin-angiotensin system, which is essential for maintaining blood pressure and body fluid balance. (P)RR contributes to the pathogenesis of various diseases, including hypertension and cancer.
When this receptor is aberrantly expressed in normal human pancreatic cell lines, genomic instability (alterations in genes and chromosomes) can occur. Such increased receptor expression contributes to the early carcinogenesis of pancreatic ductal adenocarcinoma (PDAC), the most common pancreatic cancer.
Researchers have reported that the antibodies against two (P)RR regions, located in the extracellular domain, could reduce the proliferation of human PDAC cells. Although these regions are likely to participate in cell proliferation pathway, their functional significance remains unclear.
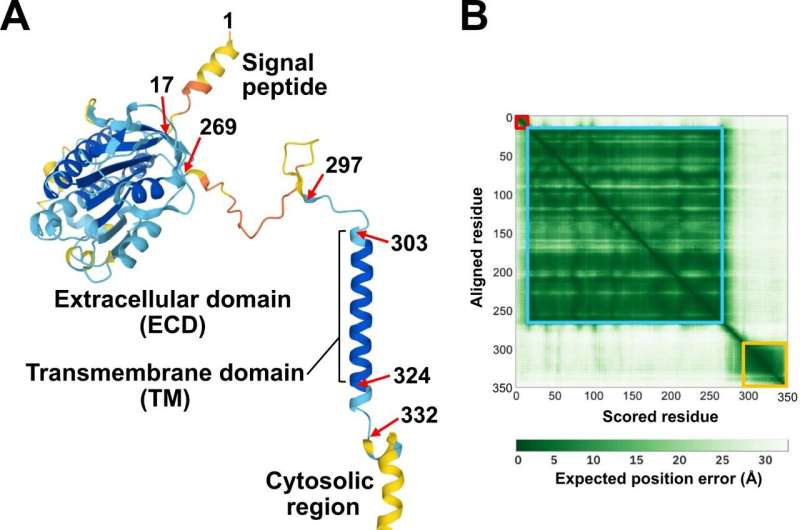
Proteins are chains of amino acids. Each protein adopts a unique 3D shape based on its amino acid sequence (the order of amino acid residues). Functionally essential amino acid residues are evolutionarily conserved and clustered to form functional patches on the protein surface. To date, the 3D structure of the (P)RR extracellular domain has not been experimentally determined.
In 2021, significant progress in protein 3D structure prediction was made using AlphaFold2 and RoseTTAFold, in which a protein 3D structural model was generated using machine learning algorithms with amino acid sequences as the only input. Both programs can predict protein structures with near-experimental accuracy.
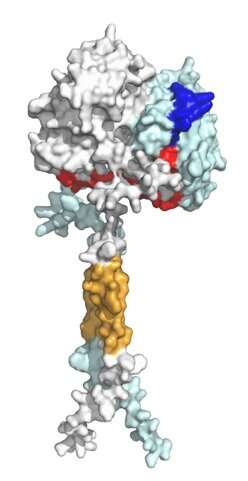
The researchers analyzed the 3D structure of (P)RR in silico using the AlphaFold2 program and evolutionary sequence conservation profile, and investigated the functional significance of the two regions involved in the PDAC antiproliferative effect.
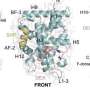
Researchers have visualized the residue positions and evolutionary sequence conservation profiles simultaneously on the 3D structure of (P)RR. The two regions mapped onto the structural model formed a continuous surface patch comprising evolutionarily conserved hydrophobic residues.
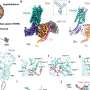
Previous reports have demonstrated that the receptor forms a dimer, meaning that the two protein units are bound together to form one assembly unit. The generated AlphaFold2 model showed that (P)RR forms a back-to-back dimer via the extracellular domain, which explains the experimentally proven dimerization.
The dimer model possessed two hand-shaped grooves with two regions of interest in the palms and an intrinsically disordered region in the fingers. Generally, an intrinsically disordered region adopts various conformations (3D shapes) under physiological conditions and can change the conformation to bind multiple partners. The surfaces of the grooves were hydrophobic, which allows for low stereospecific protein binding.
An open question regarding (P)RR functionality is its "promiscuity," meaning that beyond blood pressure regulation, the receptor can work on a wide range of molecules. Thus, it is conceivable that (P)RR utilizes its "hand" to catch two binding partners in a promiscuous manner and tether them closely in space to facilitate protein-protein interactions.
(P)RR, in particular, is thought to catch and tether two proteins involved in Wnt/β-catenin signaling, a cell proliferation pathway linked to the pancreatic cancer development. Overall, (P)RR functions as a scaffold protein, a hub for controlling the spatial and temporal organization of molecules within a cell, and thereby the flow of cellular information.
The findings of this study will be valuable for understanding disease pathogenesis and exploring novel modalities to treat human diseases, including hypertension and cancer. Further analyses are required to experimentally characterize the interactions between (P)RR and its interaction partners.
The study is published in the journal Hypertension Research.
More information: Akio Ebihara et al, Mapping the protein binding site of the (pro)renin receptor using in silico 3D structural analysis, Hypertension Research (2022). DOI: 10.1038/s41440-022-01094-w
Provided by Gifu University